Typeerror: the Dtype [] Is Not Supported for Parsing Read Csv Pandas
Pandas read_csv() tricks yous should know to speed up your data analysis
Some of the nearly helpful Pandas tricks to speed upwardly your information analysis
![]()

Importing data is the first step in whatsoever information science projection. Frequently, yous'll work with data in CSV files and run into problems at the very beginning. In this article, you'll come across how to use the Pandas read_csv() function to deal with the following common problems.
- Dealing with different grapheme encodings
- Dealing with headers
- Dealing with columns
- Parsing date columns
- Setting information type for columns
- Finding and locating invalid value
- Appending data to an existing CSV file
- Loading a huge CSV file with
chunksize
Please cheque out my Github repo for the source lawmaking.
ane. Dealing with different character encodings
Character encodings are specific sets of rules for mapping from raw binary byte strings to characters that make up the human-readable text [ane]. Python has congenital-in support for a list of standard encodings.
Character e north coding mismatches are less common today as UTF-8 is the standard text encoding in most of the programming languages including Python. However, it is definitely even so a problem if y'all are trying to read a file with a different encoding than the 1 it was originally written. Y'all are most likely to end up with something like below or DecodeError when that happens:
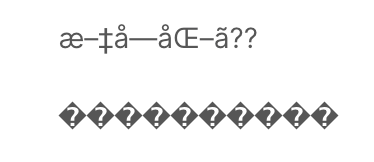
The Pandas read_csv() function has an argument phone call encoding that allows y'all to specify an encoding to use when reading a file.
Let'southward take a look at an example beneath:
First, we create a DataFrame with some Chinese characters and relieve it with encoding='gb2312' .
df = pd.DataFrame({'name': '一 二 三 四'.split(), 'northward': [2, 0, 2, 3]}) df.to_csv('data/data_1.csv', encoding='gb2312', alphabetize=False)
Then, y'all should get an UnicodeDecodeError when trying to read the file with the default utf8 encoding.
# Read it with default encoding='utf8'
# You should get an error
pd.read_csv('data/data_1.csv') 
In club to read it correctly, you should laissez passer the encoding that the file was written.
pd.read_csv('data/data_1.csv', encoding='gb2312') 
2. Dealing with headers
Headers refer to the column names. For some datasets, the headers may exist completely missing, or you might want to consider a different row equally headers. The read_csv() function has an argument called header that allows y'all to specify the headers to use.
No headers
If your CSV file does not have headers, then you need to prepare the argument header to None and the Pandas will generate some integer values every bit headers
For instance to import data_2_no_headers.csv
pd.read_csv('data/data_2_no_headers.csv', header=None) 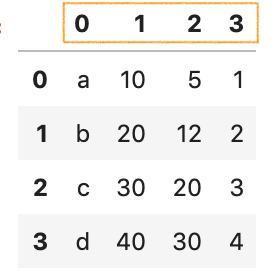
Consider different row equally headers
Allow's have a look at data_2.csv
x1, x2, x3, x4
product, price, toll, profit
a, 10, 5, 1
b, 20, 12, 2
c, thirty, xx, three
d, 40, 30, 4 It seems like more than sensible columns name would exist product, toll, … profit, but they are not in the kickoff row. The argument header as well allows you to specify the row number to use as the column names and the get-go of information. In this case, we would like to skip the offset row and use the 2nd row as headers:
pd.read_csv('data/data_2.csv', header=ane) three. Dealing with columns
When your input dataset contains a large number of columns, and yous want to load a subset of those columns into a DataFrame, then usecols will be very useful.
Functioning-wise, it is amend because instead of loading an entire DataFrame into memory and and so deleting the spare columns, nosotros can select the columns we need while loading the dataset.
Let's use the same dataset data_2.csv and select the product and cost columns.
pd.read_csv('data/data_2.csv',
header=one,
usecols=['product', 'cost']) 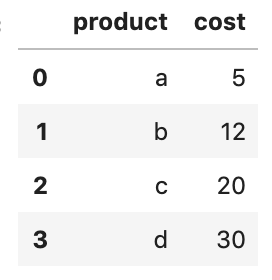
Nosotros tin can besides pass the cavalcade alphabetize to usecols:
pd.read_csv('data/data_2.csv',
header=1,
usecols=[0, 1]) 4. Parsing appointment columns
Date columns are represented as objects by default when loading information from a CSV file.
df = pd.read_csv('information/data_3.csv')
df.info() RangeIndex: four entries, 0 to 3
Data columns (full 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 engagement four non-zilch object
i product 4 non-zippo object
two toll iv non-nil int64
3 cost four not-naught int64
iv profit four non-naught int64
dtypes: int64(3), object(2)
memory usage: 288.0+ bytes
To read the engagement column correctly, we tin can use the statement parse_dates to specify a list of date columns.
df = pd.read_csv('data/data_3.csv', parse_dates=['engagement'])
df.info() <class 'pandas.core.frame.DataFrame'>
RangeIndex: four entries, 0 to three
Information columns (full five columns):
# Cavalcade Non-Null Count Dtype
--- ------ -------------- -----
0 date 4 non-zip datetime64[ns]
ane product 4 not-null object
2 toll iv non-null int64
3 price 4 not-null int64
iv profit iv not-goose egg int64
dtypes: datetime64[ns](ane), int64(iii), object(one)
memory usage: 288.0+ bytes
One-time engagement is divide into multiple columns, for example, year , calendar month , and twenty-four hour period . To combine them into a datetime, we can pass a nested list to parse_dates.
df = pd.read_csv('data/data_4.csv',
parse_dates=[['twelvemonth', 'month', 'mean solar day']])
df.info() RangeIndex: iv entries, 0 to 3
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 year_month_day 4 not-null datetime64[ns]
1 product iv non-zip object
ii price 4 non-nix int64
iii cost four non-null int64
4 turn a profit 4 non-cypher int64
dtypes: datetime64[ns](1), int64(three), object(1)
retention usage: 288.0+ bytes
To specify a custom column proper name instead of the auto-generated year_month_day , we can pass a dictionary instead.
df = pd.read_csv('data/data_4.csv',
parse_dates={ 'date': ['twelvemonth', 'month', '24-hour interval'] })
df.info() If your appointment column is in a different format, then you can customize a date parser and pass it to the argument date_parser:
from datetime import datetime
custom_date_parser = lambda x: datetime.strptime(x, "%Y %thousand %d %H:%Thousand:%S") pd.read_csv('data/data_6.csv',
parse_dates=['date'],
date_parser=custom_date_parser)
For more well-nigh parsing date columns, please check out this article
5. Setting data type
If you desire to prepare the data type for the DataFrame columns, yous tin use the argument dtype , for instance
pd.read_csv('data/data_7.csv',
dtype={
'Name': str,
'Grade': int
}) 6. Finding and locating invalid values
You might get the TypeError when setting data type using the argument dtype
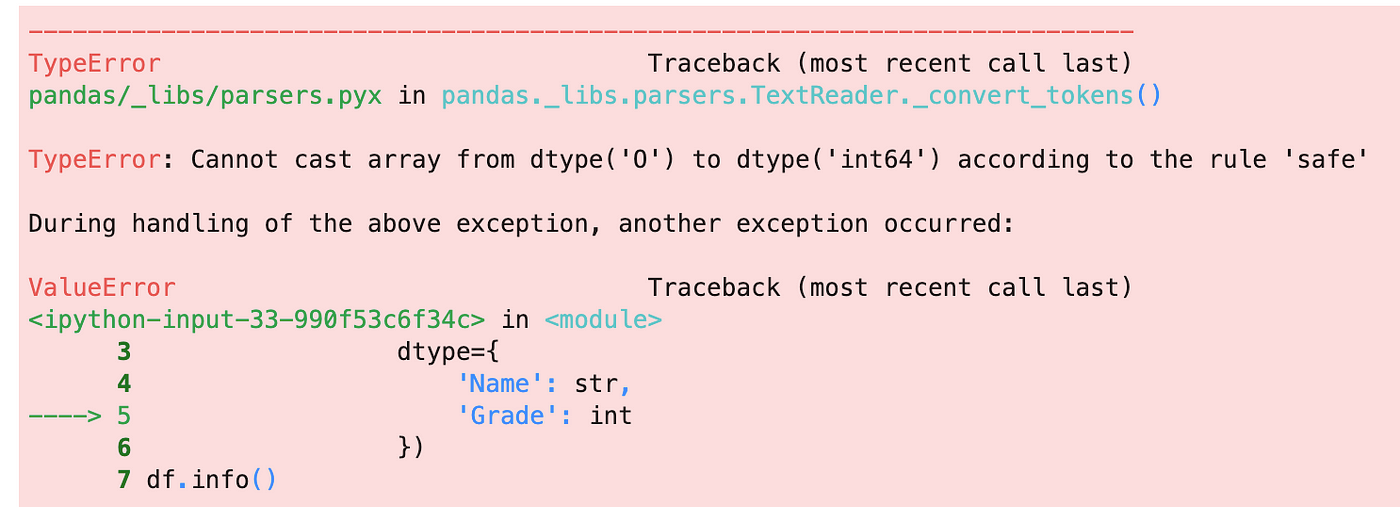
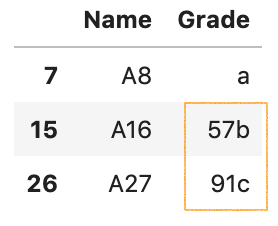
It is always useful to detect and locate the invalid values when this mistake happens. Here is how you lot can find them:
df = pd.read_csv('data/data_8.csv') is_error = pd.to_numeric(df['Grade'], errors='coerce').isna() df[is_error]
7. Appending data to an existing CSV file
Yous tin can specify a Python write mode in the Pandas to_csv() office. For appending data to an existing CSV file, we can use mode='a':
new_record = pd.DataFrame([['New name', pd.to_datetime('today')]],
columns=['Name', 'Date']) new_record.to_csv('data/existing_data.csv',
mode='a',
header=None,
index=False)
8. Loading a huge CSV file with chunksize
By default, Pandas read_csv() function will load the entire dataset into memory, and this could be a memory and performance issue when importing a huge CSV file.
read_csv() has an argument called chunksize that allows you to call back the data in a same-sized chunk. This is especially useful when reading a huge dataset as part of your information science projection.
Let's accept a look at an example below:
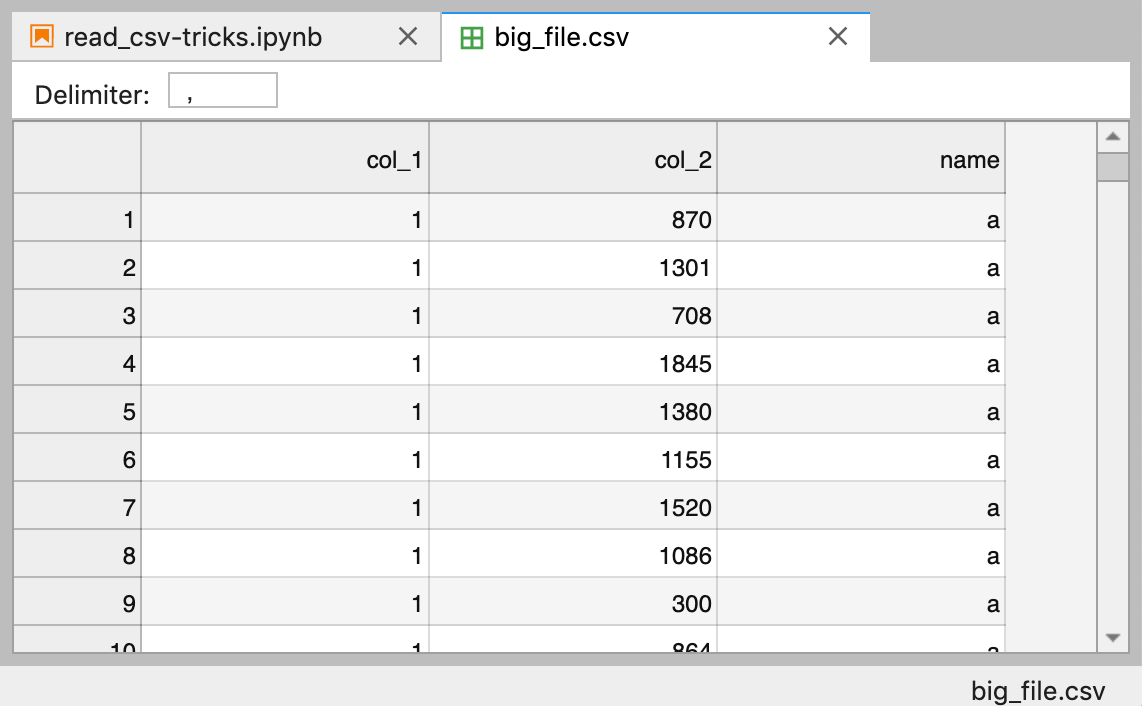
First, let'due south brand a huge dataset with 400,000 rows and save it to big_file.csv
# Make up a huge dataset nums = 100_000 for name in 'a b c d'.split():
df = pd.DataFrame({
'col_1': [i]*nums,
'col_2': np.random.randint(100, 2000, size=nums)
}) df['name'] = proper name
df.to_csv('data/big_file.csv',
fashion='a',
alphabetize=False,
header= name=='a')
Next, permit'southward specify a chucksize of l,000 when loading data with read_csv()
dfs = pd.read_csv('data/big_file.csv',
chunksize=50_000,
dtype={
'col_1': int,
'col_2': int,
'name': str
}) Let's perform some aggregations on each chunk and then concatenate the event into a single DataFrame.
res_dfs = []
for chunk in dfs:
res = chunk.groupby('proper name').col_2.agg(['count', 'sum'])
res_dfs.append(res) pd.concat(res_dfs).groupby(level=0).sum()
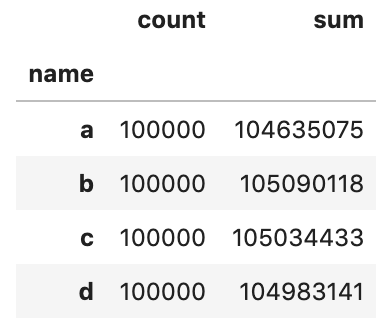
Let'due south validate the result against a solution without chunksize
pd.read_csv('data/big_file.csv',
dtype={
'col_1': int,
'col_2': int,
'proper noun': str
}).groupby('name').col_2.agg(['count', 'sum']) And you should go the same output.

That'southward it
Thank you for reading.
Please checkout the notebook on my Github for the source lawmaking.
Stay tuned if you are interested in the applied aspect of machine learning.
Here are some related manufactures
- 4 tricks you should know to parse date columns with Pandas read_csv()
- 6 Pandas tricks you lot should know to speed upward your data analysis
- 7 setups you should include at the start of a data science project.
References
- [1] Kaggle Data Cleaning: Character Encoding
Source: https://towardsdatascience.com/all-the-pandas-read-csv-you-should-know-to-speed-up-your-data-analysis-1e16fe1039f3
0 Response to "Typeerror: the Dtype [] Is Not Supported for Parsing Read Csv Pandas"
Post a Comment